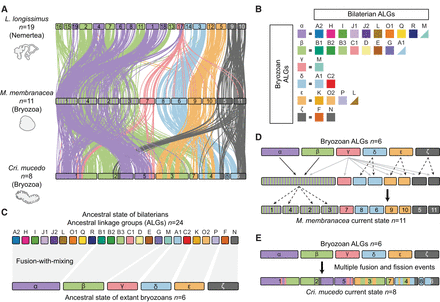
Reconstruction of ancestral genome architecture of extant bryozoans. (A) Chromosome-scale gene linkage between L. longissimus, M. membranacea, and C. mucedo. Links between orthologs are colored by the six bryozoan ALGs. Bryozoan ALGs are inferred from sets of genes that are colocated on the same chromosome (region) in both gymnolaemate and phylactolaemate bryozoans. L. longissimus is used as an outgroup because it largely preserves the 24 bryozoan ALGs on separate chromosomes but possesses the four fusion events that are shared by lophotrochozoans (J2 ⊗ L, O1 ⊗ R, O2 ⊗ K, and H ⊗ Q). (B) Composition of bryozoan ALGs by bilaterian ALGs, alongside the reconstructed bryozoan ancestral state (six chromosomes: α, β, γ, δ, ε, and ζ). Triangles indicate the inclusion of several genes from the bilaterian ALG, but the majority of genes from that ALG are elsewhere. (C) Reconstruction of the genome rearrangements that formed the bryozoan ancestral state. The top row shows bilaterian ALGs, and the bottom row shows bryozoan ALGs. (D) Reconstruction of the genome rearrangements that led from the bryozoan ancestral state to the extant gymnolaemate genomes. Solid black arrows indicate fusion-with-mixing. Solid gray arrows indicate dispersal around the genome. Dashed arrows indicate chromosome fission. (E) Reconstruction of the genome rearrangements that led from the bryozoan ancestral state to the extant phylactolaemate genomes.











