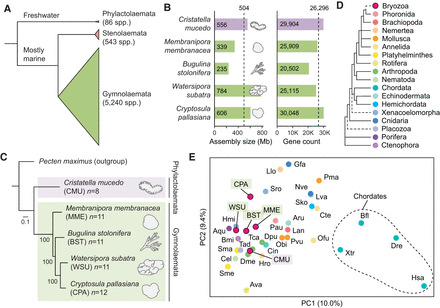
Comparative genomics of five chromosome-scale bryozoan genomes. (A) Relationships between the three classes of bryozoans. Class Phylactolaemata is the most early branching, containing just 86 species, all of which inhabit freshwater environments (Bock and Gordon 2013). Stenolaemata comprises 543 species, all of which are marine. Gymnolaemata is the most species-rich class, comprising 5240 species. (B) Genome size and gene model count of bryozoan genome assemblies used in this study. Dashed lines mark mean values. The clade containing W. subatra and C. pallasiana has larger genomes. (C) Tree of bryozoan species constructed with chromosome-scale genomes with the scallop Pecten maximus as the outgroup. P. maximus was selected as the outgroup owing to its conserved genome structure and slowly evolving sequences. The tree was constructed using the maximum likelihood method (LG + F + R6 model) with 1000 bootstrap replicates in IQ-TREE, utilizing 994 orthologous genes. The scale bar represents the number of substitutions per site. (D) Phylogenetic relationships among animal phyla, with dashed lines indicating areas of uncertainty in their placement. Phylogeny based on work of Telford et al. (2015) and Liao et al. (2023). (E) Principal component analysis (PCA) conducted using orthologous gene group counts as determined by OrthoFinder. Points are colored by their phylum, as indicated in panel D. PCs 1 and 2 explain 10.0% and 9.4% of the variance in the data set, respectively. Variance ratios for the top 30 principal components are available as Supplemental Figure S2. Species abbreviations: (Ava) Adineta vaga, (Aqu) Amphimedon queenslandica, (Aru) Asterias rubens, (Bfl) Branchiostoma floridae, (Bmi) Bolinopsis microptera, (Cel) Caenorhabditis elegans, (Cin) Ciona intestinalis, (Cte) Capitella teleta, (Dme) Drosophila melanogaster, (Dpu) Daphnia pulex, (Dre) Danio rerio, (Gfa) Galaxea fascicularis, (Hmi) Hofstenia miamia, (Hro) Helobdella robusta, (Hsa) Homo sapiens, (Lan) Lingula anatina, (Llo) Lineus longissimus, (Lva) Lytechinus variegatus, (Nve) Nematostella vectensis, (Obi) Octopus bimaculoides, (Ofu) Owenia fusiformis, (Pau) Phoronis australis, (Pma) Pecten maximus, (Pvu) Patella vulgata, (Sko) Saccoglossus kowalevskii, (Sma) Schistosoma mansoni, (Sme) Schmidtea mediterranea, (Sro) Symsagittifera roscoffensis, (Tad) Trichoplax adhaerens, (Tca) Tribolium castaneum, and (Xtr) Xenopus tropicalis.











