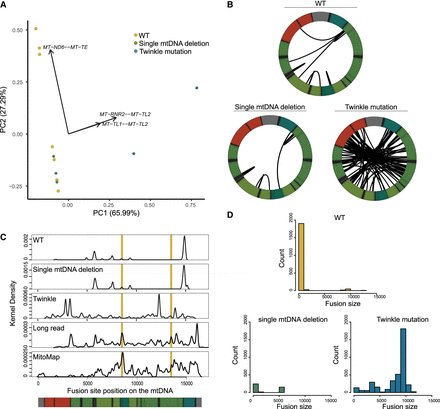
Chimeric mitochondrial RNAs (mtRNAs) in patients with mitochondrial genetic diseases. (A) Principal component analysis of human chimeric mtRNA data. (B) Distribution of chimeric mtRNAs detected more than seven times in patients with single large deletions, patients with heterogeneous mtDNA deletions owing to nuclear gene Twinkle mutations, and control subjects. (C) Location of fusion sites across the human mitochondrial genome and comparison to known deletion breakpoint distributions. Vertical yellow bars denote the location of the human 4977 or “common” deletion. (D) Histogram of fusion event sizes of chimeric mtRNAs from patients with single large deletions, Twinkle patients, and control subjects.











