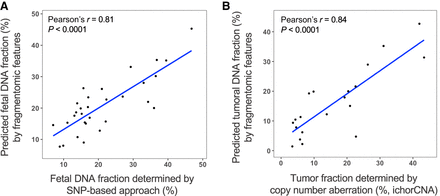
Application of fragmentomic pattern–based deduction of the fractional DNA concentration from specific cell types. Fragmentomic features, including size profile and end motif distribution, were used to train SVR models in the prediction of the fractional DNA percentage, which was correlated to the proportional tissue DNA contribution. (A) Correlation between the fetal DNA fraction predicted by the fragmentomic features and SNP-based methods, using 30 pregnant subjects. (B) Correlation between the tumoral fraction predicted by fragmentomic features and copy number aberration (ichorCNA).











