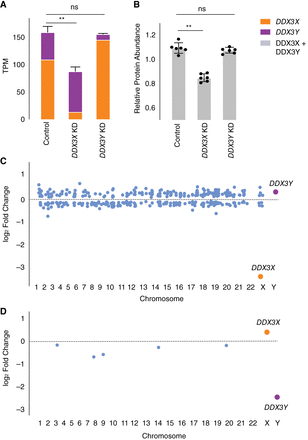
Increased expression of DDX3X fully compensates, at transcript and protein levels, for CRISPRi knockdown of DDX3Y, but the inverse is not true. (A) Stacked bar graph showing summed TPM of DDX3X and DDX3Y transcripts in knockdowns using two independent gRNAs in three independent 46,XY fibroblast cultures. Statistical significance calculated by ANOVA: (**) P < 0.001. (B) Bar graph showing abundance of shared DDX3X and DDX3Y peptides in CRISPRi knockdowns with three technical replicates in two independent 46,XY fibroblast cultures. (C) Differential gene expression analysis of control versus DDX3X knockdown reveals significant expression changes in 397 target genes across the genome, including DDX3X. Genes with P < 0.05 (after multiple hypothesis correction) are indicated in blue, with exception of DDX3X (orange) and DDX3Y (purple). (D) Differential gene expression analysis of control versus DDX3Y knockdown reveals only six genes, including DDX3Y, that change significantly.











