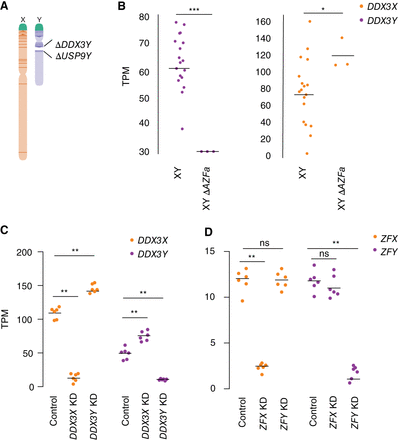
DDX3X and DDX3Y each respond to perturbations in the other's expression. (A) Schematic diagram of naturally occurring Chr Y (AZFa) microdeletion of DDX3Y and USP9Y. (B) DDX3X transcript levels are significantly higher in AZFa-deleted 46,XY LCLs compared with 46,XY LCLs with intact Chr Y. Each point represents a sample from one individual. Statistical significance determined by Mann–Whitney U-test: (***) P < 0.0001, (*) P < 0.05 (C) CRISPRi-mediated knockdown of DDX3Y using two independent gRNAs in three unrelated 46,XY fibroblast cultures results in significantly elevated DDX3X transcript levels. Conversely, DDX3X knockdown results in significantly elevated DDX3Y transcript levels. (D) Reanalysis of CRISPRi knockdown of ZFX or ZFY (San Roman et al. 2024) demonstrates that knockdown of either gene does not result in significant elevation of the homolog's transcripts. Statistical significance determined by ANOVA: (**) P < 0.001.











