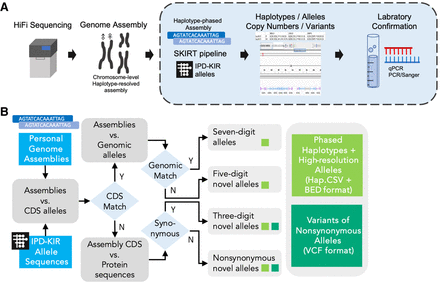
KIR allele annotation of the diploid genome assemblies. (A) Overview of the high-resolution allelic KIR identification workflow. The segment shaded blue denotes the primary focus of this study. We developed a specialized pipeline, the structural KIR annoTator (SKIRT), to facilitate annotation of high-resolution KIR alleles and structural variations in human genome assemblies. A series of laboratory experiments were conducted to validate the computational findings and ensure their accuracy. (B) The operational flow of the SKIRT pipeline. This pipeline merges a long sequence alignment tool, minimap2 (Li 2018), with in-house Python and Shell scripts. These scripts map the KIR Repository of Immuno Polymorphism Database (IPD-KIR) allele sequences against genome assemblies, such as the HPRC Year-1 release data, to identify existing KIR genes on each contig with an accuracy of up to seven-digit high-resolution alleles.











