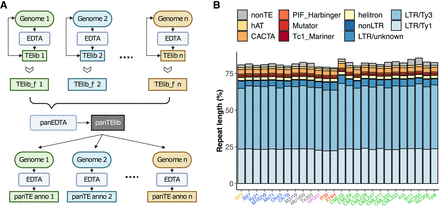
Pangenome annotation of 26 maize NAM founders using panEDTA. (A) The panEDTA workflow. The EDTA pipeline is used to annotate each genome independently, and the resulting individual TE libraries are filtered based on copy number and combined to form a nonredundant pan-TE library, which is used to reannotate each genome for a consistent pangenome TE annotation. (B) panEDTA annotation of 26 maize NAM founders. Maize lines were grouped into stiff-stalk (yellow), non-stiff-stalk (dark blue), popcorn (pink), sweet corn (red), admixed maize (gray), and tropical maize (green). Panel A was created with BioRender (https://www.biorender.com).











