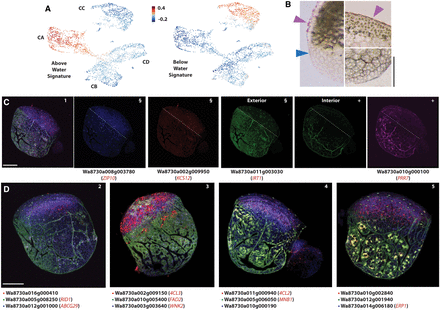
Tissue type–specific responses to life in air versus water are validated by PHYTOMap multiplex in situ RNA detection. (A) UMAP projections of “gene set activity” (Seurat) profiles of above- and below-water enriched DEGs as determined by bulk RNA-seq show the response to submerge is primarily detected in the epidermis. (B) Histological sections of Wolffia stained with Sudan IV show stained cuticle (purple arrowhead) on the exterior of the above-water epidermis (top) that is lacking from the below-water epidermis (bottom). Blue arrowhead indicates the approximate position of the waterline on the flank of the plant. (C) Select supercluster markers imaged using PHYTOMap show strong cell type specificity in line with scRNA-seq predictions. Four genes are imaged in the same plant in four fluorescence channels. From left to right: combined expression profiles; images in the blue, red, green, and magenta channels. White dashed lines indicate the approximate water line; (§/+) identical exterior or interior sections across different imaging channels. (D) PHYTOMap images showing the combined expression profiles of the genes indicated below. Composite images are a projection of 13–15 z-stack sections of 2–3 µm each. Information on genes examined can be found in Supplemental Table S2 and Supplemental Figure S2. Scale bar, 250 µm. When available, names of Arabidopsis orthologs are given (see Supplemental Data Set S1).











