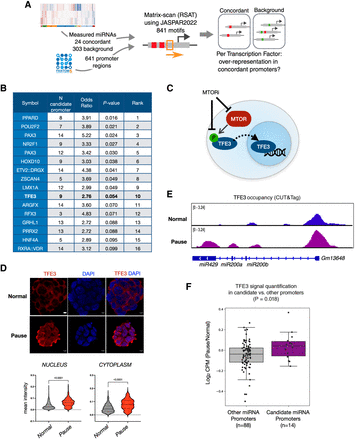
The MTOR–TFE3 axis regulates miRNA biogenesis in dormancy. (A) Schematics of TF mining at candidate miRNA promoters. Transcription start sites of miRNAs were retrieved from the FANTOM5 database and used to scan the [−1500, +500] regions with JASPAR motifs. High-confidence hits were then used to compare the fractions of promoters with a given motif, for promoters of concordant, positive logFC miRNAs against promoters of other measured miRNAs. (B) Enriched TFs at candidate miRNA promoters. TFE3, shown in bold, was chosen for experimental validation. (C) Schematics of the MTOR–TFE3 axis. MTOR phosphorylates TFE3 when active, which results in its sequestration in the cytoplasm. When MTOR is inactive, nonphosphorylated TFE3 instead translocates into the nucleus for regulation of target genes. (D) TFE3 staining in wild-type normal and paused ESCs. Bottom panels show single-cell quantifications of mean fluorescence intensity in the nucleus and cytoplasm. Scale bar, 5 µm. Statistical test is a Kolmogorov–Smirnov test, two-sided. (E) Genome browser view of TFE3 occupancy over the Gm13648 gene, which contains the miR200a, miR200b, and miR429 miRNAs. TFE3 occupancy was mapped via CUT&Tag in wild-type normal and paused ESCs. (F) Comparison of the log2 ratio of TFE3 levels (maximum scaled CPM) at candidate (n = 14) and control (n = 88) miRNA promoters. Statistical test is a Mann–Whitney U test, one-sided.











