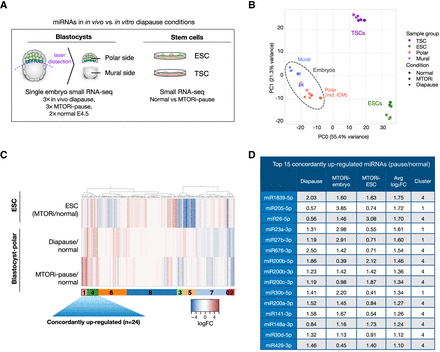
Up-regulation of a set of miRNAs is associated with dormancy. (A) Schematics of sample preparation for small RNA profiling. In vivo diapaused, in vitro diapaused, and normal E4.5 mouse blastocysts were dissected via laser microdissection to separate polar and mural ends (see Supplemental Fig. S3A). ESCs and TSCs were cultured under standard conditions with or without MTORi. Small RNA expression profiles of single embryo parts and bulk stem cells were generated via low-input small RNA-seq. (B) PCA plot of small RNA-seq data sets. Stem cells and embryos are separated along PC1 (55% variance). The polar embryo, which includes pluripotent cells, clusters closer to ESCs, whereas the mural embryo clusters closer to TSCs (PC2, 21% variance), suggesting small RNA expression profiles reflective of tissue of origin. In vivo diapaused embryos show higher variability than other groups. (C) Heatmap showing miRNA expression changes in diapaused embryos (in vitro and in vivo) compared with normal blastocysts and in paused ESCs compared with normal ESCs. miRNAs were clustered into nine clusters based on their expression levels in the three samples (for corresponding silhouette, see Supplemental Fig. S3E). Clusters 4 and 1 show a concordant trend of up-regulation on average and are the focus of the rest of the study. (D) The top 15 concordantly up-regulated miRNAs and their log2FC values in each condition.











