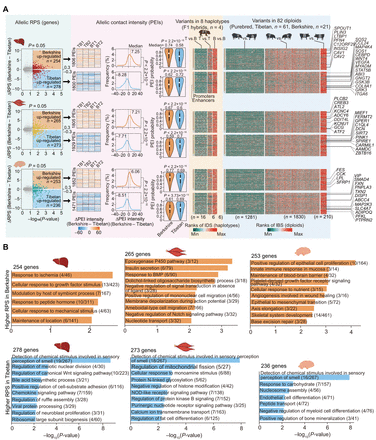
Allelic differences of PEIs in F1 hybrids. (A, left) Volcano plots of genes with a differential RPS (|ΔRPS| > 0.3 and P < 0.05, paired Student's t-test) between Berkshire and Tibetan alleles in three tissues. (Middle) Differential PEI intensities, distributions of differential PEI intensities, and estimated probabilities of PEIs based on their sequence variants (see Supplemental Methods) are shown. (Right) Degree of sequence similarity for promoters and enhancers in pairwise comparisons among eight haplotypes (Tibetan [n = 4] and Berkshire [n = 4]; measured by IDS) and among 82 diploid genomes in population-level analysis of purebred pigs (measured by pairwise IBS distances; see Supplemental Methods). Among the 82 diploids,12 from the six trios in this study (Berkshire, n = 6; Tibetan, n = 6) and 70 are publicly available (Berkshire, n = 15; Tibetan, n = 55); SNVs were retrieved from the ISwine database (http://iswine.iomics.pro/pig-iqgs/iqgs/index). Representative functional genes are labeled. (B) Significantly enriched GO-BP terms or KEGG pathways (performed using Metascape) for genes showing differential RPS between Berkshire and Tibetan haplotypes in the liver (left), muscle (middle), and brain (right).











