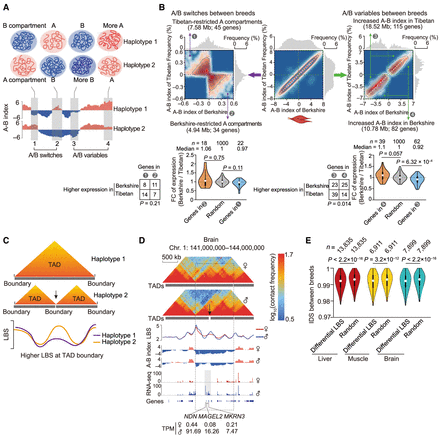
Compartmental rearrangements and variable TAD boundaries among haploid genomes. (A) Identification of compartmental rearrangements. Regions with distinct compartment status were defined (i.e., from B [negative value of A-B index] to A [positive value of A-B index], or from A to B; scenarios 1 and 2, respectively) as A/B switched regions. Additionally, compartment scores (i.e., the A-B index values) for regions with the same status were compared between haploid genomes (not between those with distinct switches in compartment status) and identified regions with significantly higher compartment scores as A/B variable regions (P < 0.05, paired Student's t-test, and |ΔA-B index| > 0.5; scenarios 3 and 4). (B) Allelic compartmental rearrangements in muscle. We identified compartment A regions exclusively found in the Tibetan (7.58 Mb) or Berkshire (4.94 Mb) alleles (top left) and regions with significantly elevated compartment scores in the Tibetan (18.52 Mb) and Berkshire (10.78 Mb) alleles (top right). Genes with testable differences in allele (i.e., with informative SNVs and TPM ≥ 0.5 in at least one allele) tended to be located in the more accessible chromatin regions and generally showed increased expression; violin plots show changes in allelic expression (Wilcoxon rank-sum test) and counts (Fisher's exact test) of genes located in regions with allelic differences in compartmentalization (i.e., A/B switches, left; A/B variables, right). (C) Identification of shifted TAD boundaries (see Supplemental Methods). (D) Paternal-specific TAD boundaries adjacent to three paternally expressed imprinted genes (NDN, MAGEL2, and MKRN3). (E) In comparison to genomic background, regions with allelic differences in boundary strength (LBS, P < 0.05, paired Student's t-test) showed greater allelic divergence in sequence divergence (reflected by the lower pairwise haplotype similarities between two parental breeds determined by identity score [IDS]) (see Supplemental Methods). P-values are from a Wilcoxon rank-sum test. Hi-C maps at 20-kb resolution were used to generate these graphs.











