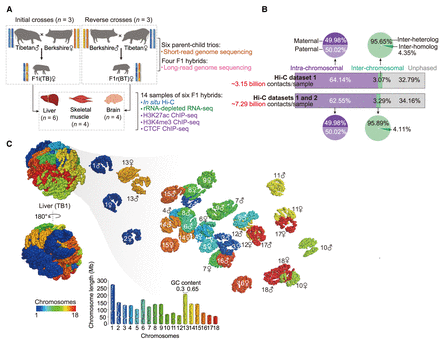
Three-dimensional (3D) structures of diploid genomes in hybrid pigs. (A) Schematic representation of reciprocal crosses between genetically distinct Berkshire and Tibetan pig breeds. Liver, skeletal muscle, and brain of F1 hybrids were collected for multi-omic assays (for details, see Supplemental Fig. S1). F1i and F1r denote the F1 hybrids of initial and reverse crosses, respectively. (B) Allele assignment of in situ Hi-C contacts in F1 hybrids. A total of about 48.95 billion valid Hi-C contacts were generated, including about 3.15 billion autosomal contacts for each of the 14 samples (six liver, four muscle, and four brain samples; data set 1). To reliably identify PEIs throughout the diploid genome, we performed additional in situ Hi-C assays for 12 samples (data set 2), resulting in a final total of about 97.53 billion valid Hi-C contacts (about 7.29 billion autosomal contacts for each of the 12 samples, including four liver, four muscle, and four brain samples) in the combined data sets 1 and 2 (for details, see Supplemental Fig. S1). (C) 3D genome structure of a representative liver sample (see Supplemental Methods). (Left) Whole genome; (right) the 18 autosome pairs visualized separately.











