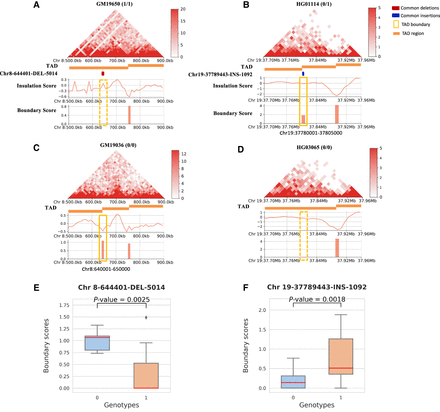
Visualization of two SVs that disrupt TAD boundaries with significant changes in boundary strength. (A,C) A deletion (Chr 8-644401-DEL-5014) that disrupts the TAD boundary and shows differences in Hi-C contact maps, BSs, and insulation scores for individuals with (genotype 1/1) and without (genotype 0/0) the deletion. The orange rectangle shows the TAD location for each sample within the plot region. The dark red rectangle represents the location of this deletion, and the yellow rectangle highlights the TAD boundary location and corresponding boundary strength. The top left figure is the GM19650 sample, whose genotype is 1/1, i.e., it carries this deletion, compared to the sample below, GM19036, whose genotype is 0/0, i.e., it does not have this deletion. The BS panel shows that the GM19650 sample lacks the TAD boundary where it carries that genomic deletion. (B,D) Similar comparisons for an example of an insertion (Chr 19-37789443-INS-1092) between the individual HG01114 (genotype 0/1) and HG03065 (genotype 0/0). The BS panel shows that the HG01114 sample has the TAD boundary where it carries that genomic insertion. (E,F) Boxplots demonstrating the significant differences in BSs for two genotype categories, 0 (genotype 0/0) and 1 (genotypes 0/1, 1/0, or 1/1).











