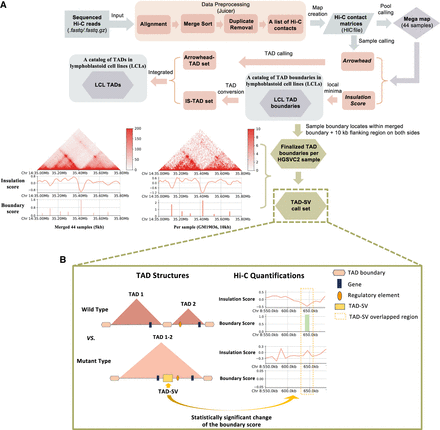
The step-by-step workflow to process raw Hi-C data into TADs/TAD boundaries and TAD–SVs in our Hi-C analysis pipeline. (A) The raw read files from 44 samples were used as input in Juicer for preprocessing and generating Hi-C maps, which were subsequently binned at multiple resolutions. The Insulation Score (IS) algorithm was applied to call an initial TAD boundary for each sample. All 44 Hi-C libraries were merged together to create a “mega” map and used as an input of Arrowhead (Rao et al. 2014) and IS (Crane et al. 2015) algorithms to call TADs, and TAD boundaries for the LCL merged call set. Finalized TAD boundary results for each individual were defined as those sample boundaries located within the merged boundary plus 10 kb flanking regions (the size of the exact TAD boundary called by IS for each individual) on the left side of the boundary start site and the right side of the boundary end site (Yu et al. 2017). The two figures located in the bottom left corner are shown as a comparison between the merged subject level and single subject level, which includes the Hi-C contact maps, the insulation scores, and the boundary strengths for the merged call set (5 kb) and the GM19036 (10 kb) sample over the region Chr 14: 35–35.8 Mb. (B) We examined the impact of SVs on chromatin structure by measuring the boundary score for each TAD boundary with the presence or absence of SVs. The Wilcoxon rank-sum test was employed to identify SVs significantly affecting TAD boundary strength, resulting in a set of TAD–SVs.











