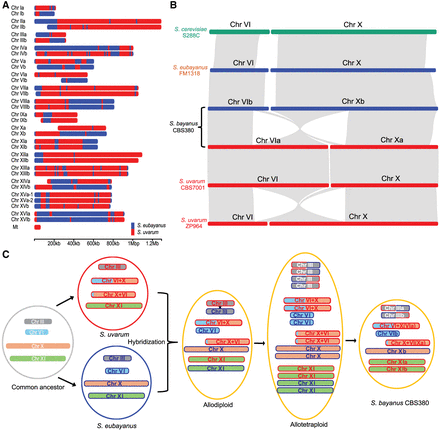
The origin and evolution of S. bayanus chromosomes. (A) Origin of genomic regions of each chromosome in the S. bayanus genome based on BLAST searches of nonoverlapping 5000 bp blocks. Genomic regions that originated from S. eubayanus are shown in blue, and regions inherited from S. uvarum are shown in red. (B) Synteny block of Chr VI and Chr X between S. cerevisiae, S. eubayanus, both S. bayanus haplotypes, S. uvarum strain CBS7001, and S. uvarum strain ZP964. (C) An evolutionary model of S. bayanus chromosomes. For simplification purposes, only four chromosomes are shown, representing different patterns of chromosome inheritances. Translocation between Chr VI and Chr X occurred in S. eubayanus prior to its hybridization with S. bayanus. Recombination and whole-genome duplication occurred in the hybrid S. bayanus genome. Subsequent genome reduction, probably by sporulation, created some heterozygous chromosomes, such as Chr III, and some homozygous chromosomes, such as Chr XI.











