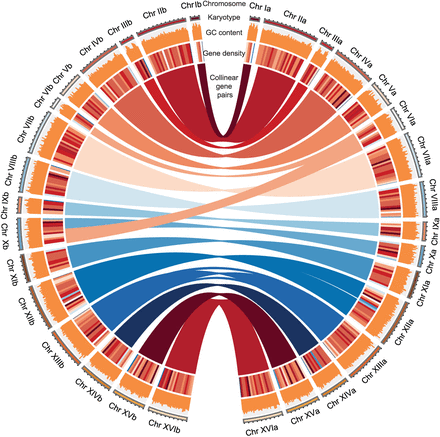
Circos circular visualization of the genome assembly for the 16 pairs of chromosomes of the S. bayanus genome. Different genomic features across four concentric circles. The outermost circle represents the karyotype of the S. bayanus genome for two haplotypes, with the right part representing haplotype-a and the left part representing haplotype-b. The second outermost circle represents the GC content on each chromosome of the genome. The third circle provides information on gene density within the chromosomes, and a darker color indicates a higher gene density. The innermost circle highlights synteny blocks between haplotypes, illustrating collinear gene pairs between haplotype-a and haplotype-b.











