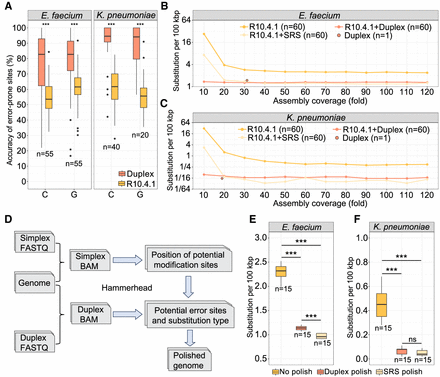
Polishing with duplex reads resolves DNA modification-induced errors in R10.4.1 assembly. (A) Read accuracy comparison for R10.4.1 and duplex reads at error-prone C and G sites. (B,C) Rates of SNS in R10.4.1 assemblies following short-read or duplex polishing in E. faecium and K. pneumoniae, respectively. The assembly only with duplex reads is also shown as a reference. (D) The workflow exclusively focuses on polishing the potential modification sites using duplex reads. (E,F) Comparison of SNS rates in R10.4.1 assemblies following targeted duplex and short-read polishing, focusing on 601 and 369 possible modification sites identified by Hammerhead in E. faecium and K. pneumoniae, respectively. (***) P-value < 1 × 10−8, Student's t-test. (SRS) short-read sequencing.











