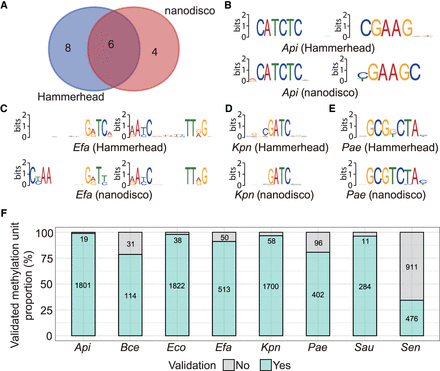
Validation of the potential modification sites from Hammerhead results. (A) The number of shared and unique motifs identified by Hammerhead and nanodisco. (B–E) The comparison of six shared motifs from two pipelines. (F) The proportion of overlap between two groups of modification units identified by Hammerhead and nanodisco. Each modification unit consists of a potential modification site and the surrounding bases (−4 bp to +4 bp). Note: Hammerhead was designed for R10.4.1 reads, while nanodisco was for R9.4.1 reads. (Api) Acinetobacter pittii, (Bce) Bacillus cereus, (Eco) Escherichia coli, (Efa) Enterococcus faecium, (Kpn) Klebsiella pneumoniae, (Pae) Pseudomonas aeruginosa, (Sau) Staphylococcus aureus, (Sen) Salmonella enterica.











