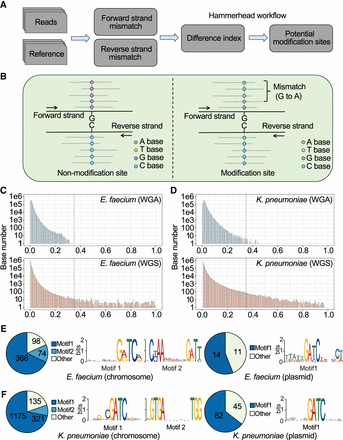
Bacterial DNA modifications can be identified by comparing the mapping accuracy between forward and reverse strands in R10.4.1 reads. (A,B) The workflow and demo of Hammerhead, which is designed to identify the potential modification sites based on the degree of nucleotide difference between forward and reverse reads. (C,D) The distribution of site difference index in WGA and WGS sequencing reads for E. faecium and K. pneumoniae, respectively. The WGA sequencing, representing random read errors, serves as a background filter. High discrepancies between forward and reverse strands in WGS reads suggest potential DNA modifications. A cutoff of 0.35 (FDR < 1 × 10−6 in WGA reads) is used here to identify possible DNA modification sites in WGS reads. (E,F) The motif was enriched from possible DNA modification sites identified by strand accuracy comparison for chromosome and plasmid sequences in E. faecium and K. pneumoniae, respectively. “GATC” is the dominating motif. Note: Only one motif was enriched from E. faecium plasmid (MEME E-value = 1.1 × 10−7). (FDR) false discovery rate.











