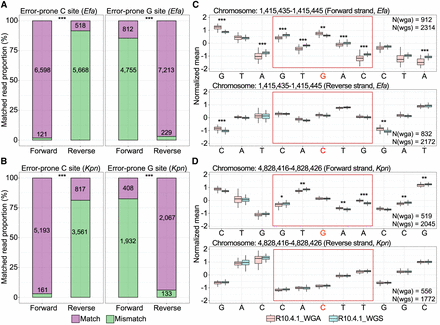
The error-prone sites arising from bacterial DNA modifications show strand bias in R10.4.1 reads. (A,B) Proportions and counts of accurately and inaccurately mapped forward and reverse read at error-prone C and G sites in E. faecium and K. pneumoniae, respectively. (***) P-values <1 × 10−30, Fisher's exact test. (C,D) Illustration of raw current signals at an error-prone G site in E. faecium and K. pneumoniae genome, respectively. Notably, significant differences highlighted between WGS and WGA reads are observed in the forward strand (upper panel), but not in the reverse strand (lower panel). (wgs) whole-genome shotgun, (wga) whole-genome amplification, (Efa) Enterococcus faecium, (Kpn) Klebsiella pneumoniae. (*) P-value < 0.05, (**) P-value < 0.01, and (***) P-value < 0.001, Student's t-test.











