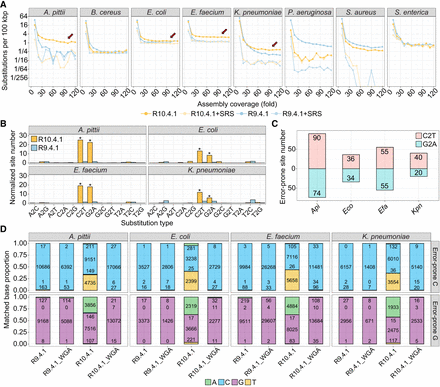
Bacterial DNA modifications influence substitutions in R10.4.1 read–based assemblies. (A) Substitution per 100 kbp of assemblies generated using different coverage of R10.4.1 and R9.4.1 reads, with or without high-quality short-read polishing. The x-axis shows the subsampled read coverage for ONT reads. (B) Normalized per-assembly counts of all 12 SNS types in A. pittii, E. faecium, E. coli, and K. pneumoniae assemblies generated from R10.4.1 (n = 35) or R9.4.1 (n = 35) reads. The R10.4.1 and R9.4.1 assemblies are based on subsampled read coverage from 40- to 100-fold. (*) P-value < 0.01, Student's t-test. (C) Identification of error-prone sites in four bacteria defined by C2T or G2A substitution frequencies exceeding two. (D) Proportions and counts of accurately and inaccurately mapped bases at error-prone C and G sites in four bacterial genomes, respectively. Mapping reads were obtained from R9.4.1, R9.4.1 WGA, R10.4.1, and R10.4.1 WGA. Notably, the C2T and G2A substitutions are more prevalent in R10.4.1 reads. (SRS) short-read sequencing, (Api) Acinetobacter pittii, (Eco) Escherichia coli, (Efa) Enterococcus faecium, (Kpn) Klebsiella pneumoniae.











