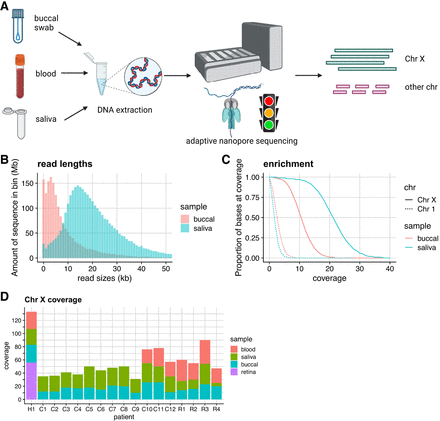
X inactivation skew sequencing workflow. (A) Adaptive nanopore sequencing of peripheral samples (blood, saliva, and buccal mucosa) to maximize coverage of the X Chromosome. Sequencing of fragments that do not map to the X Chromosome is terminated early. Degraded buccal DNA results in shorter read lengths (B) and reduced X Chromosome enrichment performance (C). The read lengths for (accepted) reads mapping to the X Chromosome are plotted for patient C9 for illustrative purposes. Rejected reads are typically <1 kb. (D) X Chromosome coverage for each patient, split by sample. At least 30× combined X Chromosome coverage was obtained for each patient. (H1) Healthy individual, (C1–C12) female carriers of choroideremia, and (R1–R4) female carriers of RPGR-associated X-linked retinitis pigmentosa.











