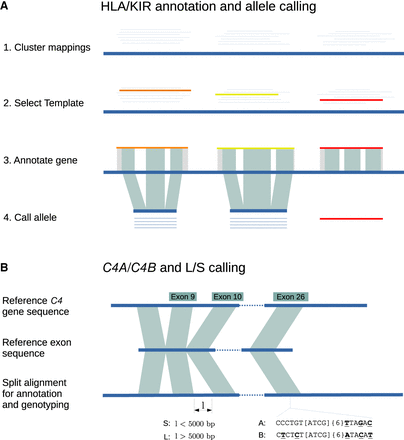
Overview of the Immuannot algorithm. (A) Annotate HLA/KIR genes using known alleles from the IPD-IMGT/HLA or the IPD-KIR databases. Gene sequences (thin line) from the IPD databases are mapped to the target contig sequence (thick line) at step 1. At step 2, the best-matching allele with the smallest number of mismatches and the longest alignment length is chosen as the template allele (in different colors) for each mapping cluster. Project the gene structure of template allele to the contig to annotate exons, introns, UTRs, CDS, and start and stop codons (step 3). At step 4, determine the allele type based on the CDS similarity. (B) Annotate the C4 genes. Align the C4 transcript sequence from GRCh38 to the contig sequence. Infer C4A versus C4B based on the sequence on exon 26 and infer the long (L) versus short (S) form based on the intron size between exons 9 and 10.











