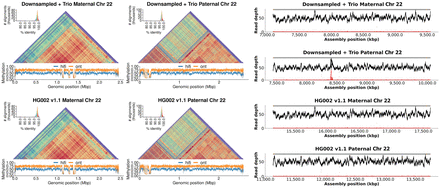
ONT-only assemblies accurately resolve centromeric arrays for both haplotypes. The figure shows StainedGlass (Vollger et al. 2022) and methylation plots for Chromosome 22 of HG002 on the left and NucFreq (Vollger et al. 2019; Mc Cartney et al. 2022) validation using HiFi sequencing (Jarvis et al. 2022; Liao et al. 2023) on the right. The top row shows the 50× Duplex + 30× UL + trio assembly while the bottom is the HG002 v1.1 reference assembly (https://github.com/marbl/HG002/blob/main/README.md). The alpha satellite repeat pattern is consistent between both assemblies for both haplotypes. The methylation pattern, including the location of the centromeric dip region (CDR) (Logsdon et al. 2021; Altemose et al. 2022; Gershman et al. 2022), is also consistent between assemblies. Lastly, NucFreq shows the assembly is overall accurate, with a few local quality issues indicated by an increase in secondary allele frequency (red), likely due to a missing centromeric repeat unit caused by the lower read accuracy.











