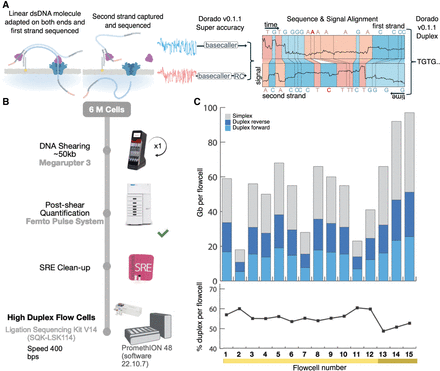
Duplex data generation. (A) Sequences with an adapter on both strands are sequenced sequentially. Once sequenced, the reads are processed using the stereo basecaller. First, each strand of the sequence is converted to basecalls using the super high-accuracy mode of the basecallers. The segmented signals and the bases of the strands are then aligned to each other and a “stereo” basecalling model is run which combines this information to give a final sequence for the double-stranded molecule. Note that the basecaller in this study was run both on the instrument to detect and call reads where both strands were sequenced as well as on the output reads marked as single-stranded to identify missed double-strand junctions. (B) The process for library preparation before sequencing. DNA is sheared to 50 kb followed by clean-up before sequencing on the PromethION. (C) The throughput and yields from the cells used for HG002 in this study. The yield in terms of total bases is indicated by the bars. After conversion to Duplex, the forward and reverse strands are combined, yielding a single read. While variable, the Duplex yield stabilized at ∼20 Gb per flow cell in the later sequencing runs with the newest flow cells (Supplemental Table 1, mustard yellow).











