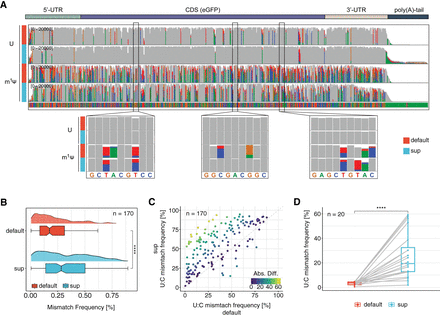
SUP improves the identification of m1Ψ-modified residues in synthetic mRNA vaccines. (A) IGV screenshot showing the synthetic mRNA vaccine containing either unmodified uridine (U) or N1-methylpseudouridine (m1Ψ). The 20,000 longest, uniquely mapped reads were selected for this analysis. Positions at which the mismatch frequency exceeds 0.2 are colored, and those with a mismatch frequency below 0.2 are shown in gray. (B) Comparison of mismatch frequency across all m1Ψ sites (n = 170) found in the synthetic eGPF vaccine. To test for statistical significance, the nonparametric Wilcoxon-test was used, and values were corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure: (ns) P > 0.05, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001. (C) Comparison of U > C mismatch frequency for all 170 m1Ψ sites between the default and SUP. Points located in the upper half of the dotted line represent sites with higher U > C frequency in SUP (n = 151), and points below the dotted line represent sites with higher U > C frequency in the default. (n = 19). Points are colored by the absolute difference between the default and SUP. (D) Comparison of U > C mismatch frequency across the most lowly modified sites in the default (median = 3.45) compared with the SUP (median = 19.43). A paired two-sided Wilcoxon test was performed to test for statistical significance. For B,D, the box is limited by the lower-quartile Q1 (bottom) and upper-quartile Q3 (top). Whiskers are defined as 1.5 × IQR with outliers removed.











