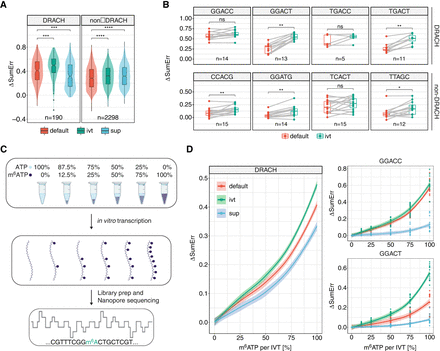
The effect of custom basecalling models on biologically relevant m6A motifs and at different m6A stoichiometries. (A) Comparison of the delta summed errors (ΔSumErr = SumErrMOD − SumErrUNM) at the central modified position for 5-mers that fall within the DRACH motif (D = A, G, or U; R = G or A; H = A, C, or U) and others (non-DRACH). Statistical analysis was performed using a two-sided nonparametric Wilcoxon test with “default” as the reference group. (B) Comparison of the ΔSumErr for individual motifs that are DRACH and non-DRACH. Each dot represents a single observation of that 5-mer within the “curlcake” sequences. To compare the relationship between individual 5-mers, a paired two-sided Wilcoxon test was performed. (C) Schematic representation of the experimental approach to obtain “curlcakes” at different m6A stoichiometries. ATP and m6ATP were mixed at different ratios and used for the in vitro transcription of “curlcakes” with the remaining NTPs, followed by poly(A)-tailing, library preparation, and direct RNA sequencing (DRS). (D) Comparison of error-signature amplitude at different modification stoichiometries between 5-mers that fall within the DRACH motif. The corresponding plot for non-DRACH motifs is reported in Supplemental Figure S5B. Lines were fitted using locally estimated scatterplot smoothing (loess), and lightly shaded areas correspond to the 95% confidence interval. For A,B the box is limited by the lower-quartile Q1 (bottom) and upper-quartile Q3 (top). Whiskers are defined as 1.5 × IQR with outliers represented as individual dots. All statistical tests were corrected for multiple hypothesis testing using the Benjamini–Hochberg correction, and adjusted P-values are displayed: (ns) P > 0.05, (*) P ≤ 0.05, (**) P ≤ 0.01.











