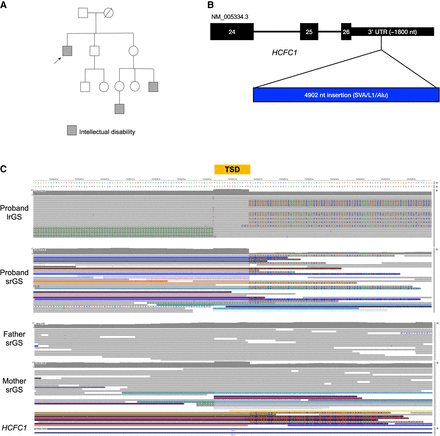
Proband 3 has a 4 kb insertion in the 3′ UTR of HCFC1. (A) The proband's family has a history of X-linked ID, as the proband (arrow) and two other male relatives (gray squares) are affected. (B) Model of the relative length of the insertion in the 3′ UTR NM_005334.3. Only exons 24–26 are shown. (C) The hemizygous insertion is likely inherited from a heterozygous carrier mother, as indicated by srGS reads. Note that some reads contain unaligned ends (multicolored bases) while some span the entire insertion (purple line at the 5′ end of the target site duplication [TSD]); all the proband's reads support the insertion. A TSD is a hallmark of an L1-mediated insertion.











