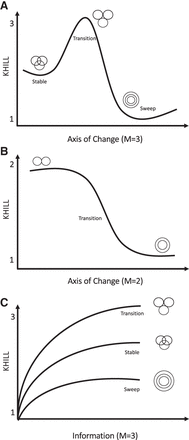
KHILL cartoons. In a population genomic context, KHILL can be used to track changing clinical genomic complexity over time and/or space. We expect that in a pandemic, a VOC will increase the information diversity of a stable population. If this transition is favored, the variant will sweep the population (A). In a pooled or wastewater context, KHILL can function as a measure of compression of unassembled, raw sequence across days, with a drastic change revealing the potential arrival of a new variant (B). Genomes sampled from a transitioning population will exhibit higher KHILL than a population that has settled into a dominant variant (C).











