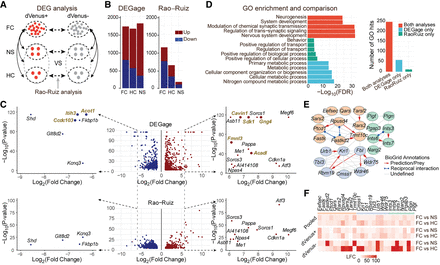
DEGage detects DEGs related to remote memory formation. (A) Organization of the analyses performed by Rao-Ruiz and DEGage. (B) Comparison of the numbers of DEGs detected by DEGage and Rao-Ruiz. (C) Volcano plots of DEGs showcase DEGs detected by DEGage and Rao-Ruiz. Subfigures highlight DEGs with log2 fold changes (LFCs) less than −6 and greater than six. DEGs uniquely identified by DEGage are marked in gold. (D) The enrichment of GO biological process of the DEGs identified by both DEGage and Rao-Ruiz. The bar plot shows the number of GO biological process hits for each of the previously listed conditions. (E) Networks retrieved from STRING, with their corresponding BioGrid annotations, derived from DEGs uniquely identified by DEGage. These genes primarily fall into three major functional groups of GO biological processes (shown in light blue, red, and green). LFCs of these genes are presented in F.











