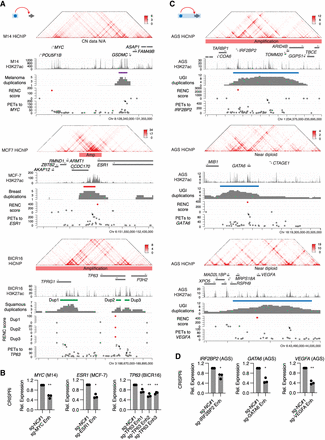
Different patterns of enhancer duplications activating cancer-related genes. (A) Duplications of noncoding enhancers activating promoters of the oncogenes MYC in melanoma, ESR1 in breast cancer, and TP63 in squamous cancer. Presented tracks include H3K27ac HiChIP signal, H3K27ac ChIP-seq signal, positions of the identified duplication hotspots, duplication events observed in the corresponding cancer types, RENC scores prioritizing gene promoters that are more likely to be activated by enhancers within the duplication hotspot, and the number of PETs of each significant interaction between the enhancers and the RENC-prioritized gene promoter. The SNP-array-based copy number (CN) status of the hotspot in the corresponding cancer cell lines is indicated (the SNP-array data are not available for the M14 cell line). Note that the RENC analysis prioritized the short isoform of TP63 (deltaNP63) as the target gene for the duplication hotspot presented in squamous cancer, so that the RENC score of the deltaNP63 promoter is used to represent the TP63 gene. (B) CRISPRi of the indicated enhancers for MYC, ESR1, and TP63 caused significant reductions in their expression, as measured by RT-qPCR, in the M14, MCF-7, and BICR16 cell lines, respectively (n = 3 biological replicates). P-values were derived from two-sided t-tests: (*) P < 0.05, (**) P < 0.01. (C) Duplications of noncoding enhancers together with their target genes such as IRF2BP2, GATA6, and VEGFA in UGI cancer. Similar to A, presented are H3K27ac HiChIP signal, H3K27ac ChIP-seq signal, positions of the duplication hotspots, the duplication occurrence within the highlighted windows, RENC scores prioritizing gene promoters that are more likely to be activated by enhancers within the hotspot, and the number of PETs of each significant interaction between the enhancers and the RENC-prioritized gene promoter. SNP-array-based copy number status of the hotspot in AGS cells is indicated. (D) CRISPRi of the indicated enhancers for IRF2BP2, GATA6, and VEGFA caused significant reductions in their expression, as measured by RT-qPCR, in AGS cells (n = 3 biological replicates). P-values were derived from two-sided t-tests: (*) P < 0.05, (**) P < 0.01.











