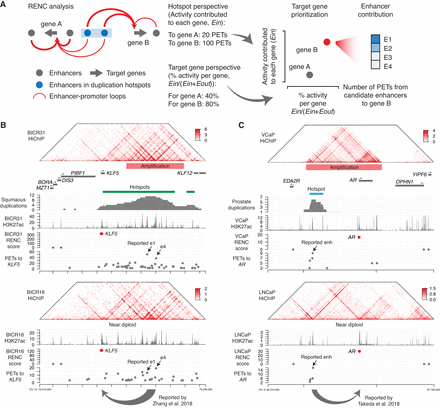
A methodology to identify the target genes activated by the duplicated enhancers. (A) Schematic illustrating the HiChIP-based RENC methodology that we applied to rank the target genes for the enhancers within the duplication hotspots. (B) Presented tracks include H3K27ac HiChIP signal, positions of duplication hotspots, duplication events observed in squamous cancer, H3K27ac ChIP-seq signal, RENC scores prioritizing the KLF5 promoter that is more likely to be activated by enhancers within the left duplication hotspot presented in the window, and the number of PETs contributed by each interacting enhancer to KLF5. (Upper) Data from BICR31 cells with the KLF5 enhancer amplification; (bottom) data from BICR16 cells without amplification at the KLF5 locus. Two of the previously published functional enhancers for KLF5 were indicated (Zhang et al. 2018). (C) Presented tracks include H3K27ac HiChIP signal, positions of duplication hotspots, duplication events observed in prostate cancer, H3K27ac ChIP-seq signal, RENC scores prioritizing the AR promoter that is more likely to be activated by enhancers within the duplication hotspot, and the number of PETs contributed by each interacting enhancer to AR. (Upper) Data from VCaP cells with AR and its enhancers amplified; (bottom) data from LNCaP cells that are near diploid at the AR locus. A previously published functional enhancer for AR was indicated (Takeda et al. 2018).











