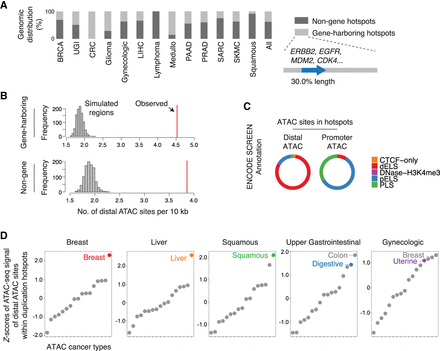
Enrichment of distal enhancers in the duplication hotspots. (A) The percentage of the identified duplication hotspots harboring intact protein-coding genes (from the start codon to the stop codon) in each cancer type. For gene-harboring hotspots, the harbored genes only account for 30.0% length of the hotspots wherein the genes are found. (B) The number of distal ATAC sites per 10 kb within the duplication hotspots or randomly shuffled regions matching the lengths of the hotspots. (C) The percentage of the distal and promoter ATAC sites residing in different categories of regulatory elements annotated in the ENCODE SCREEN data set. (dELS) distal enhancer-like signature, (pELS) proximal enhancer-like signature, (PLS) promoter-like signature. (D) Z-scores of the averaged ATAC-seq signal of distal ATAC sites within duplication hotspots (sum of ATAC signal for all distal ATAC sites within the hotspots divided by the number of ATAC-seq profiled samples) across different cancer types.











