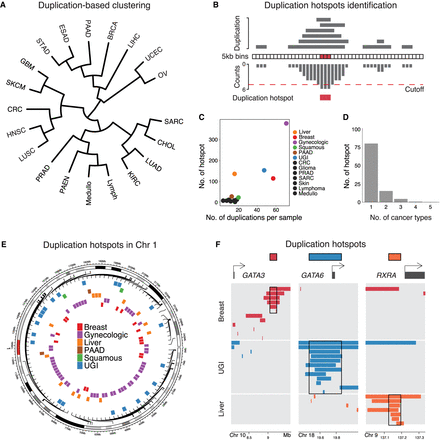
Identification of duplication hotspots in the cancer genome. (A) Hierarchical clustering of cancer types based on the duplication frequencies of continuous 5 kb bins in the genome. The full names of the cancer types are listed in Supplemental Table S1. (B) Schematic illustrating the methodology applied to identify duplication hotspots in each cancer type. Based on a Poisson distribution, 5 kb bins that are duplicated more often than expected were identified (FDR < 0.05). The significant 5 kb bins that are adjacent to each other or one 5 kb bin apart were then stitched together as a duplication hotspot. (C) Presented are the number of duplication events observed per sample (x-axis) and the number of identified duplication hotspots (y-axis) in each cancer type. (D) The number of the identified duplication hotspots that are unique to a single cancer type or shared by multiple cancer types. (E) The distribution of the identified duplication hotspots from multiple cancer types in Chromosome 1. (F) The distribution of the duplication events observed at the GATA3, GATA6, and RXRA loci in breast, upper gastrointestinal (UGI), and liver cancers.











