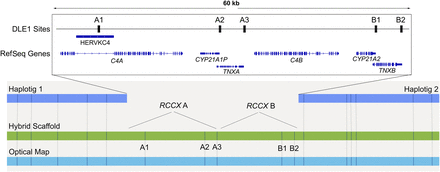
AFA RCCX evaluation using Bionano hybrid assembly. Haplotigs (dark blue) are combined with Bionano optical maps (light blue) to create a hybrid scaffold (green). The gap in between the haplotigs represents the RCCX CNV missing from the assembly. Each RCCX copy has at least two DLE-1 sites in each of the CYP21 and TNX genes, represented (A2, A3, B1, B2); A third DLE-1 site (A1) will be present if there is a HERVKC4 insertion in the C4 intron. Based on the count of the missing DLE-1 sites, the RCCX count and layout can be inferred.











