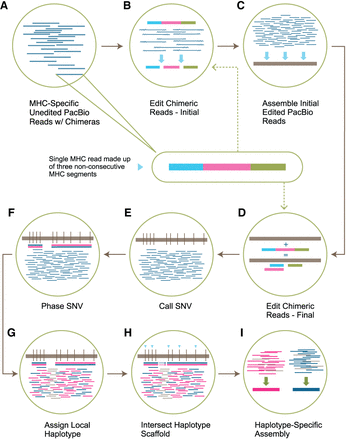
Read editing and haplotype binning detail. (A) Unedited PacBio reads that align to one of the 160 MHC haplotypes in the reference sequence. Inset shows the composition of a single read containing three nonconsecutive or palindromic MHC sequences (chimera) represented by three colors. (B) The purpose of this step is to split each chimeric read into individual segments (initial edited reads). This is done by overlapping chimeric reads and identifying positions along each chimeric read with a buildup of overlap termination sites. (C) Initial edited reads are assembled into haploid mosaic contigs. (D) A final round of chimeric read editing aims at further improving the detection of the chimera boundaries. The chimeric MHC-specific reads from A are aligned to the collapsed assembly, which acts as a reference sequence. The chimeric read is then split based on the reported aligned segments (final edited reads). (E) SNVs are called across the collapsed assembly and (F) SNVs are phased using the final edited read alignments. (G) Final edited reads are assigned a local haplotype if they intersect with variants in a phased block (pink segment = haplotype 1, blue segment = haplotype 2, gray segment = on both haplotypes). (H) Each locally phased block is oriented to the full MHC haplotype by intersecting with the reference-based haplotype scaffold (light blue triangles). (I) Haplotype-specific edited reads are combined with homozygous reads and assembled independently into haplotype-specific assemblies.











