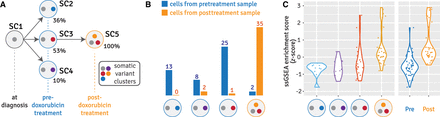
Single-cell assignment and subclone-specific EMT signature enrichment analysis in a longitudinal breast cancer data set. (A) Tumor subclone evolution reconstructed from bulk whole-genome DNA sequencing data across doxorubicin treatment of a refractory breast cancer patient from our previous study (Brady et al. 2017). (B) Cell-to-subclone assignment from scRNA-seq data collected from the tumor samples before and after doxorubicin treatment. Blue bars and numbers represent the number of cells from the pretreatment sample assigned to each subclone; orange bars and numbers represent the number of cells from the posttreatment sample assigned to each subclone. (C) Z-score of ssGSEA enrichment scores for epithelial to mesenchymal transition (EMT) signature: grouped by subclone (left), and by time point (right).











