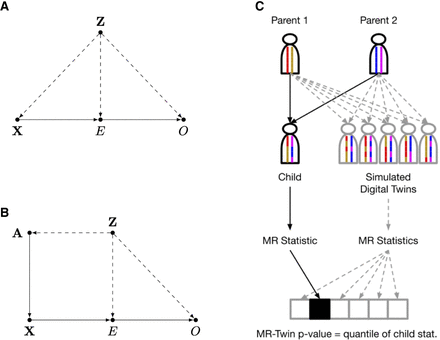
Illustrations of Mendelian randomization (MR) assumptions and the MR-Twin framework. (A) Directed acyclic graph (DAG) depicting variables and their relationships in a typical MR study, where X is the genotypic instrument, E is the exposure trait, and O is the outcome trait. An external confounder Z, such as population stratification, can cause violations of the MR assumptions. (B) If we have the parental haplotypes A, then X is independent of Z given A. (C) Illustration of the MR-Twin workflow. Digital twin genotypes are sampled from the parental genotypes. MR-Twin is a conditional randomization test, conditioned on A and therefore immune to confounding from Z, in which the P-value is computed based on the quantile of the true offspring's MR-Twin statistic compared with the digital twins’ statistics.











