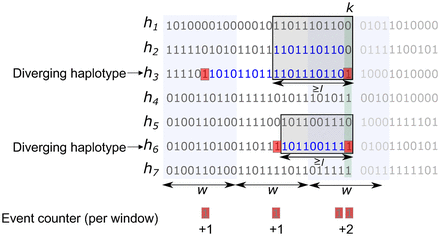
A snapshot of the FastRecomb algorithm for counting potential recombination events at the variant site k. When sweeping through the PBWT panel, haplotypes are sorted by their reverse suffix ending at site k. FastRecomb identifies diverging haplotypes, that is, haplotypes carrying minor alleles at site k in all l-blocks (boxes) ending at site k. Then, both ends (red shading) of the current match (blue) of a diverging haplotype with other haplotypes of the block will trigger a potential crossover event. All events are captured by “event counter” ϕi and ρi for each window i of length w.











