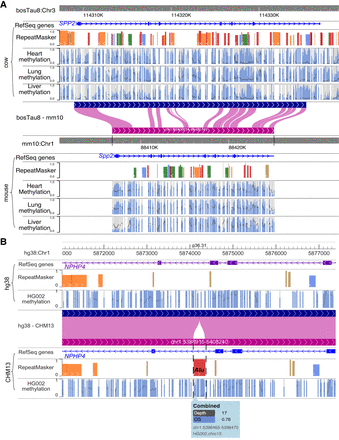
Applying comparative genomic analysis to nonmodel organisms and new genome assemblies. (A) Creating a cattle–mouse comparative browser view and using it to compare DNA methylation in the heart, lung, and liver between the cow and mouse. RefSeq genes and RepeatMasker tracks along with DNA methylation status of the heart, lung, and liver tissues from both the cow and mouse were plotted on the Comparative Epigenome Browser. (B) Using the browser to compare the difference between hg38 and CHM13 and how it may affect genomic analysis. The same HG002 WGBS data were mapped to hg38 and CHM13, respectively. The DNA methylation difference by either genome is a minimum across most of the genomic region, but an Alu insertion is only present in the CHM13 reference, and the hypermethylation of this Alu element can only be assessed using the CHM13 reference. Both methylation percentage and read coverage of each CpG site were annotated within the methylC tracks. All CpG sites are marked by gray, with methylation percentage annotated by the blue bar in the foreground (0% methylated CpGs are displayed as full gray bars, whereas 100% methylated CpGs are displayed as full blue bars). The read coverage over CpG sites across the region is represented by the black line in the background.











