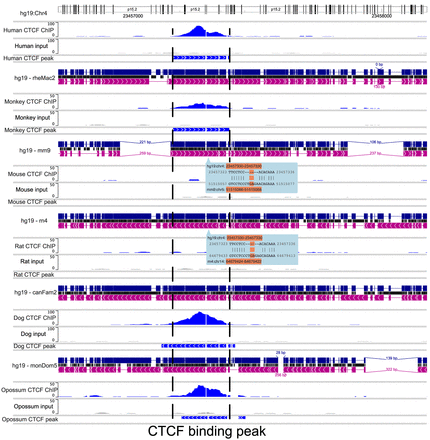
Using the WashU Comparative Epigenome Browser to visualize and compare the CTCF binding sites from six mammals. CTCF ChIP-seq and input from human (hg19), rhesus macaque (rheMac2), mouse (mm9), rat (rn4), dog (canFam2), and opossum (monDom5) were displayed on the browser. Human reference genome hg19 was used as the reference genome, and all the other species were anchored to their orthologous region from hg19 using whole-genome alignments. The region hg19:Chr4:23,456,625–23,458,090 shows a conserved CTCF binding peak in the orthologous loci in all mammal genomes except the two rodents, indicating a rodent-specific loss of a conserved CTCF binding site. The loss of CTCF binding also coincided with a rodent-specific 6-bp insertion.











