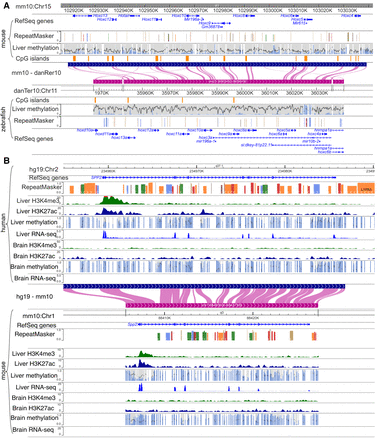
Comparing conserved epigenomic features between species. (A) The DNA methylation status of the Hoxc gene cluster is conserved between the mouse and zebrafish. Mouse and zebrafish DNA methylomes were characterized by Zhang et al. (2016). Mouse and zebrafish reference genomes (mm10 and danRer7) are shown back-to-back anchored by the mouse–zebrafish genome-align track along with their gene, repeat, and CpG island annotations. Liver DNA methylome data are from Zhang et al. (2016) with enhanced reduced representation bisulfite sequencing (ERRBS) displayed. (B) H3K4me3 and H3K27ac ChIP-seq, WGBS, and RNA-seq of brain and liver samples from both human and mouse of the SPP2/Spp2 gene are displayed using the WashU Comparative Epigenome Browser. Both DNA methylation level and read depth are illustrated in the methylC track. Both methylation percentage and read coverage of each CpG site were annotated within the methylC tracks. All CpG sites are marked by gray, with methylation percentage annotated by the blue bar in the foreground (0% methylated CpGs are displayed as full gray bars, whereas 100% methylated CpGs are displayed as full blue bars). The read coverage over CpG sites across the region is represented by the black line in the background.











