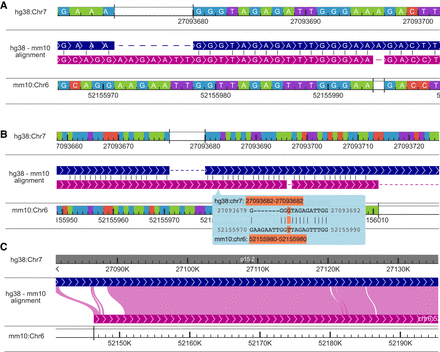
Displaying genome alignments using the WashU Comparative Epigenome Browser. (A) Displaying hg38-mm10 BLASTZ alignment at the nucleotide level with more than 10 pixels per nucleotide. The sequence strand in the alignment is illustrated using arrows. Syntenic nucleotides from hg38 and mm10 are vertically aligned with gaps inserted. Matching nucleotides are connected using a short vertical line in the alignment track. (B) Displaying hg38-mm10 alignment between 0.1 pixels per nucleotide and 10 pixels per nucleotide. The alignment is organized the same as in panel A without displaying nucleotides within the alignment. Alignments at nucleotide resolution are visible in the cursor tip hover box, and the nucleotide alignment under the cursor is highlighted in orange (G–T). (C) Displaying alignment with >10 nt per pixel. Both hg38 and mm10 genomes are continuously displayed without breaks. Syntenic regions are connected using pink Bezier curves.











