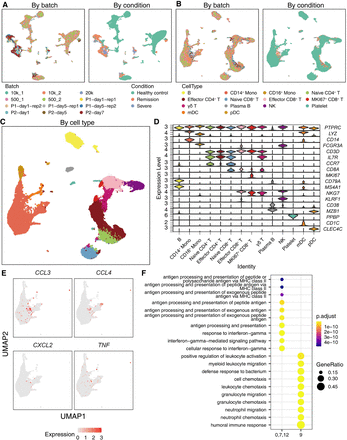
scInt reveals inflammatory-related mechanisms in the COVID-19 data. (A,B) UMAP visualizations of the COVID-19 PBMC data in the raw data (A) and scInt-integrated data (B). Cells are colored by batches and COVID-19 stage conditions. (C) UMAP visualizations of the scInt-integrated data. Cells are colored by identified cell types. (D) Stacked violin plot of the top-important marker gene expression for each cell type. (E) Overlay of the expression patterns of the cytokine storm-related and inflammation-related chemokine genes in the CD14+ monocytes. (F) The top 10 enriched GO biological processes associated with cluster 9 and other CD14+ monocytes.











