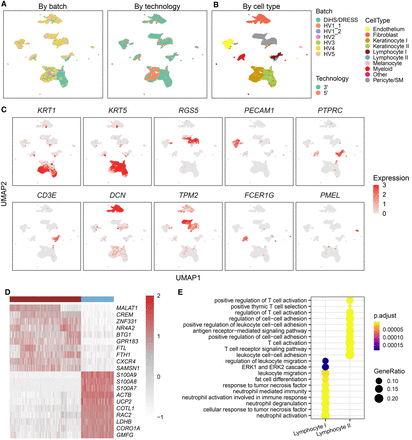
scInt reveals DiHS/DRESS-specific cell subpopulations. (A,B) UMAP visualizations of the human DiHS/DRESS and healthy skin cells in the scInt-integrated data. Cells are colored by batches, technologies (A), and annotated cell labels (B) determined in the scInt-integrated data by examining the expression patterns of known markers. (C) Overlay of the expression patterns of cell type marker genes onto the UMAP space. (D) Heatmap of the top 10 differentially expressed genes of the DiHS/DRESS and healthy lymphocyte subpopulations. (E) The top 10 enriched GO biological processes associated with the DiHS/DRESS and healthy lymphocyte subpopulations.











