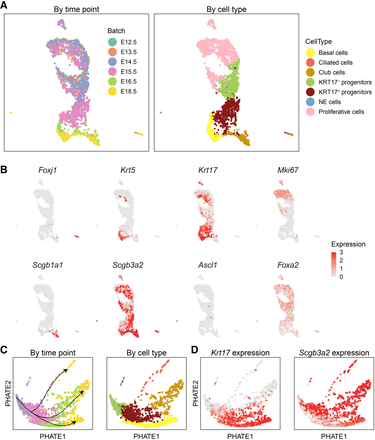
scInt reveals integrated trajectories of mouse developing tracheal epithelial data. (A) UMAP visualizations of the mouse developing tracheal epithelial cells in the scInt-integrated data (from E12.5 to E18.5). Cells are colored by time points (left) and cell types (right). (B) Overlay of the expression patterns of cell type marker genes onto the UMAP space. (C) PHATE visualizations for all cells except proliferative cells. Cells are colored by time points (left) and cell types (right). The arrows indicate the directions of the trajectories. (D) Overlay of the expression patterns of Krt17 and Scgb3a2 onto the PHATE space.











