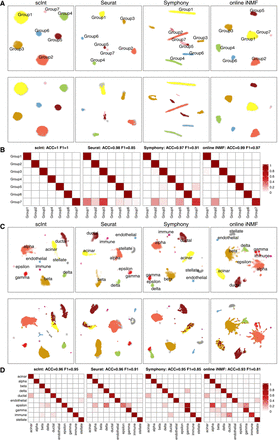
Benchmarking scInt against other methods in the reference-based integration tasks. (A) UMAP visualizations of the density of integrated reference cells (top row) and the scatters of mapped query cells (bottom row) of simulation 2, from scInt, Seurat, Symphony, and online iNMF. Cells are colored by ground-truth cell type labels, with gray shadows representing the reference. (B) Heatmap comparing the 5-NN predicted labels (columns) and the original labels (rows) of the query. The color bar indicates the proportion of query cells per original cell type label that was predicted to be for each reference label (rows sum to 1). (C) UMAP visualizations of the density of integrated reference cells (top row) and the scatters of mapped query cells (bottom row) of the human pancreas data, from scInt, Seurat, Symphony, and online iNMF. Cells are colored by ground-truth cell type labels, with gray shadows representing the reference. The mast cells are the unique cell type in the query data and are colored dark purple. (D) Heatmap comparing the 5-NN predicted labels (columns) and the original labels (rows) of the query.











