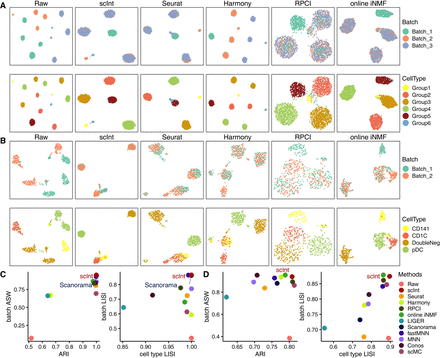
Benchmarking scInt against other methods using simulated and real data. (A) UMAP visualizations of simulation 1 before and after integration by scInt, Seurat, Harmony, RPCI, and online iNMF. Cells are colored by batch labels (top row) and cell type labels (bottom row). (B) UMAP visualizations of the human dendritic data before and after integration by scInt, Seurat, Harmony, RPCI, and online iNMF. (C) The comparison of evaluation metrics, including ARI, batch ASW, cell type LISI, and batch LISI, of the integrated results on simulation 1. (D) The comparison of evaluation metrics, including ARI, batch ASW, cell type LISI, and batch LISI, of the integrated results on human dendritic data.











